Atlas
Julich Brain Atlas
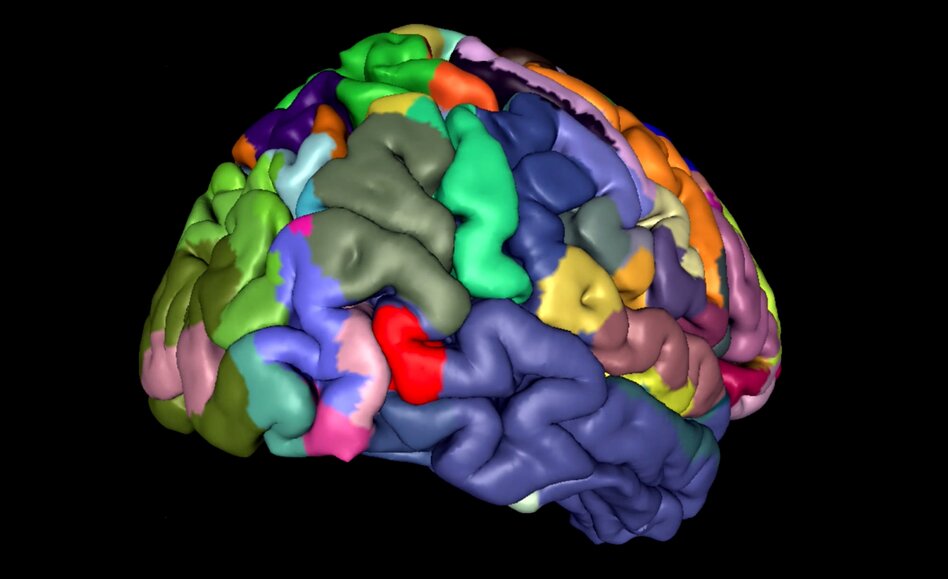

Mapping the human brain at multiple scales
The Julich Brain Atlas framework is a three-dimensional atlas of the human brain which replaces historical maps of the brain's microstructure such as that of Brodmann. It integrates high-resolution cytoarchitectonic maps with microstructural and connectivity data as well as neurotransmitter receptor expression profiles and functional data – all into a common reference space.
It is continuously expanded and openly accessible for researchers to systematically integrate multi-level data. It is interoperable in a way that it can be linked with other brain parcellations, databases and mapping projects. The Julich Brain Atlas offers a powerful tool for neuroscience and medicine alike.
Atlas Elements
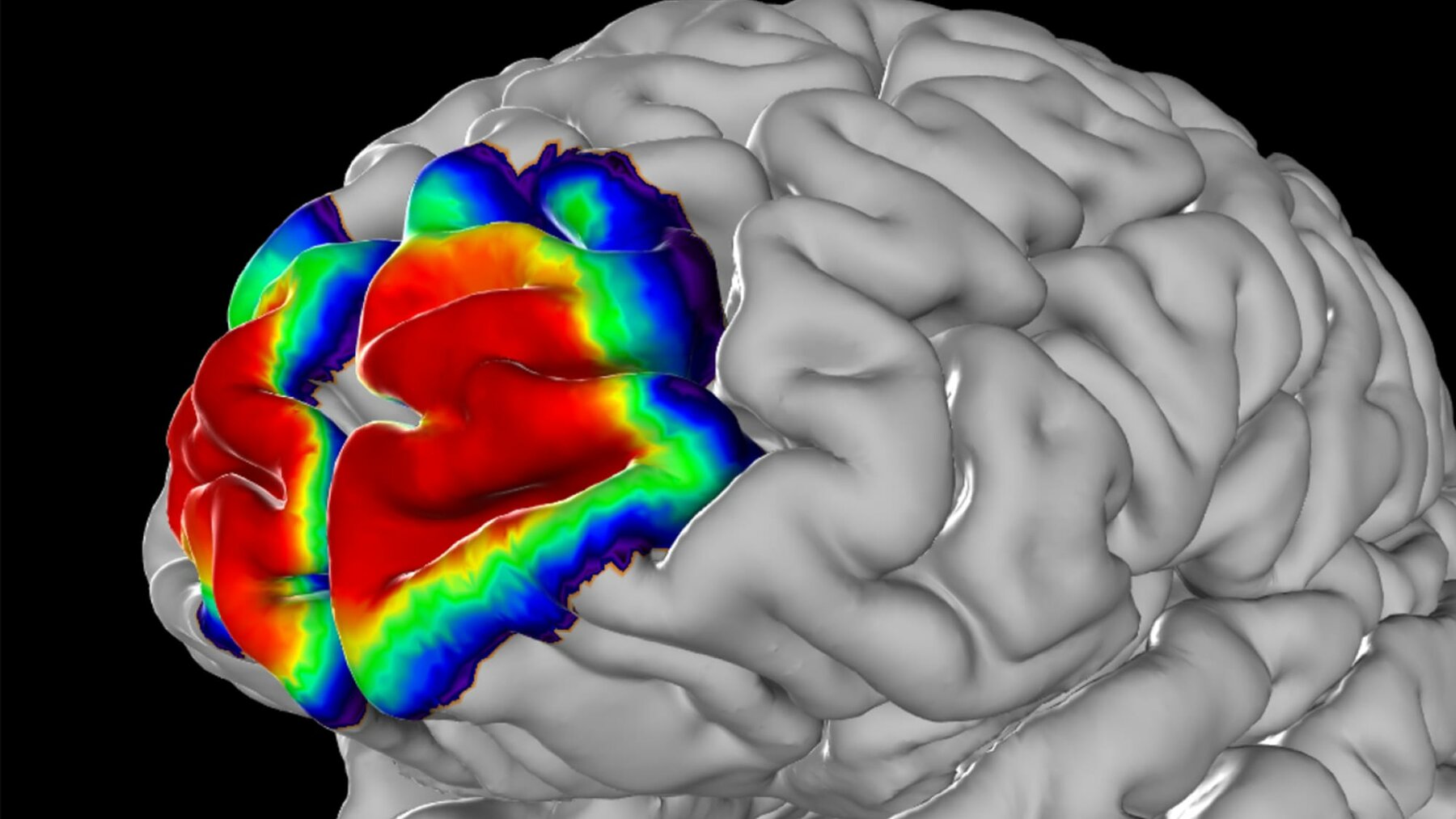
Probabilistic Maps
3D probabilistic maps of cytoarchitectonic regions
Tap to learn more
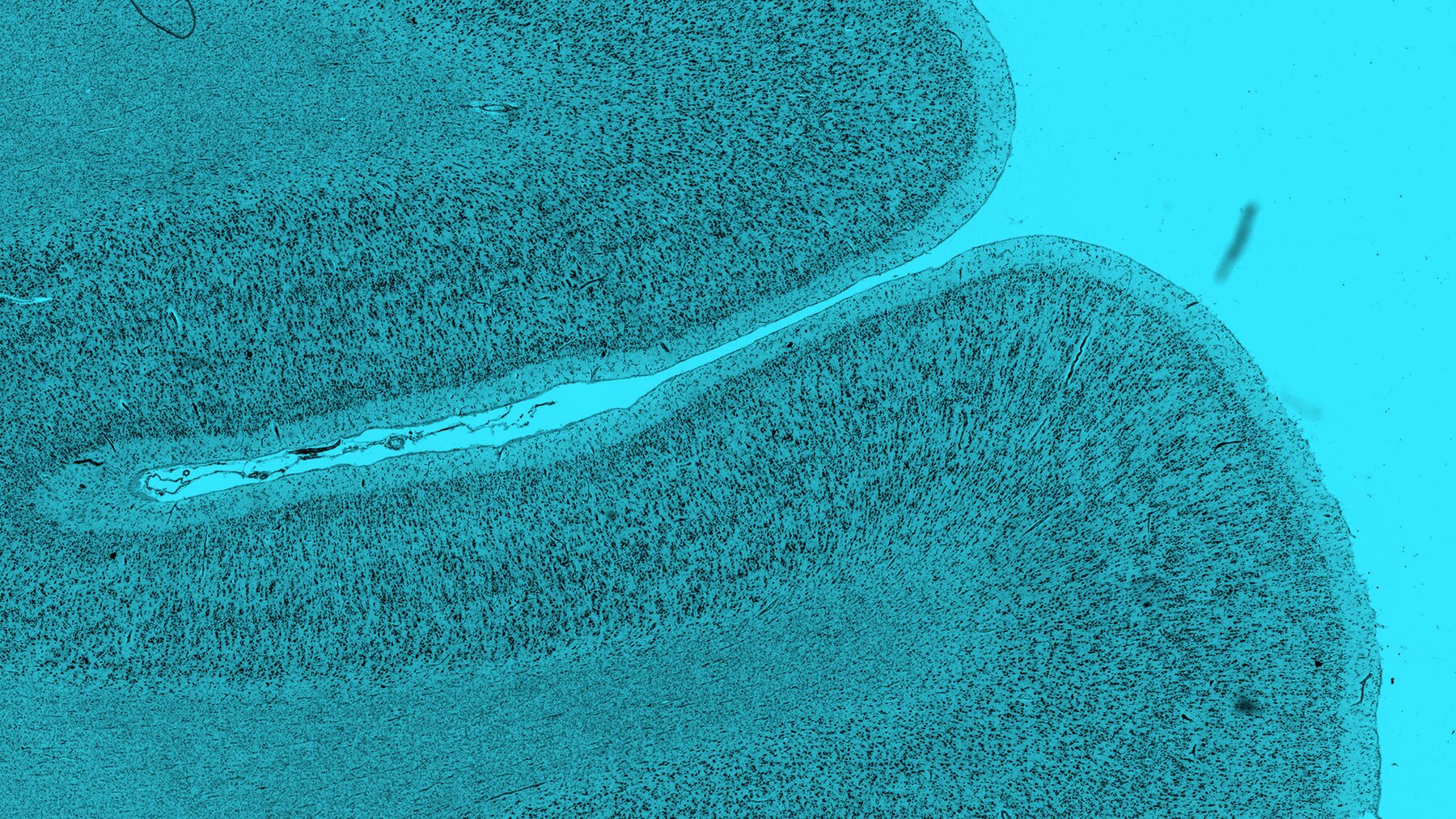
BigBrain
Reference brain in 20 micrometer resolution
Tap to learn more
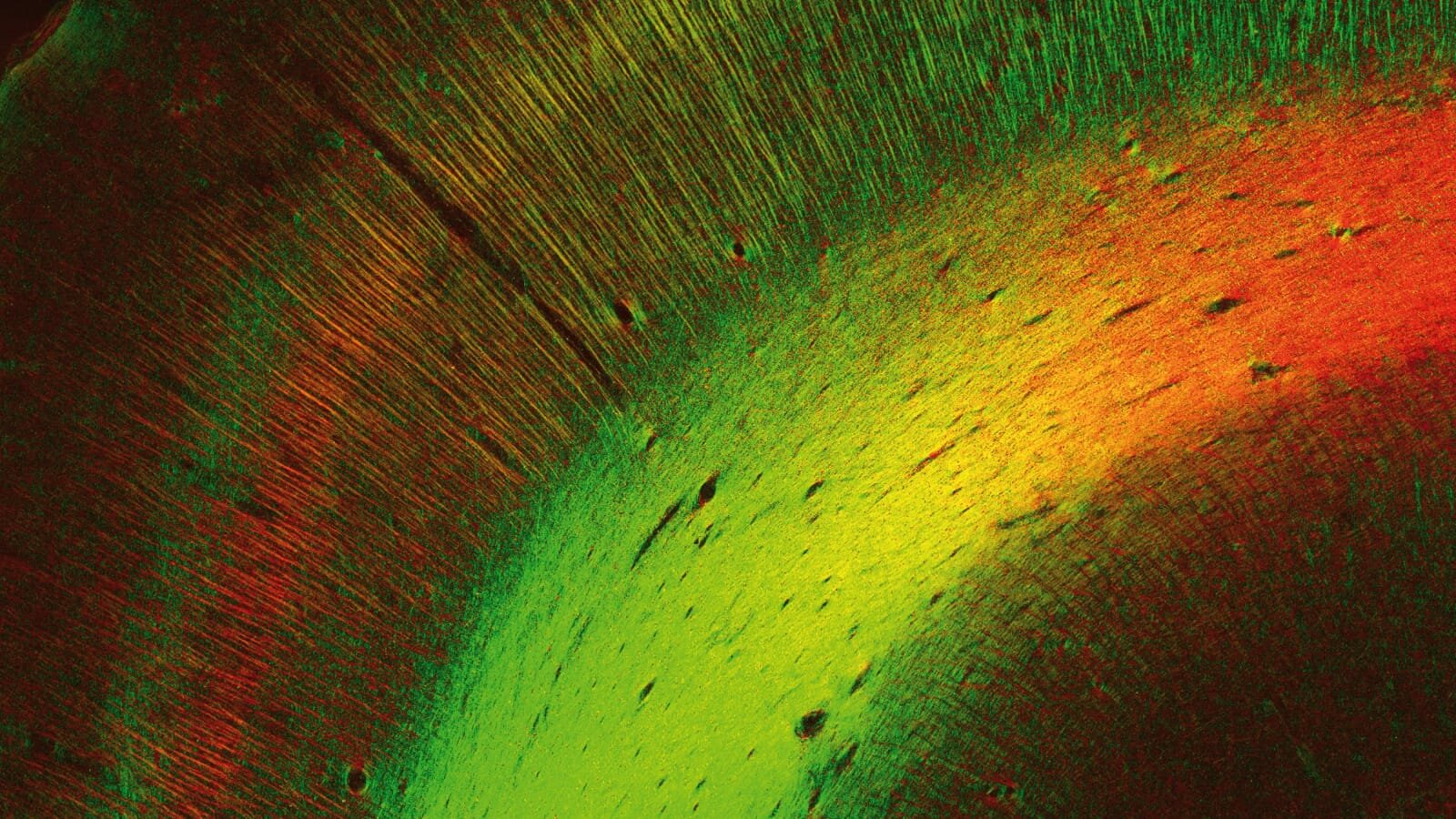
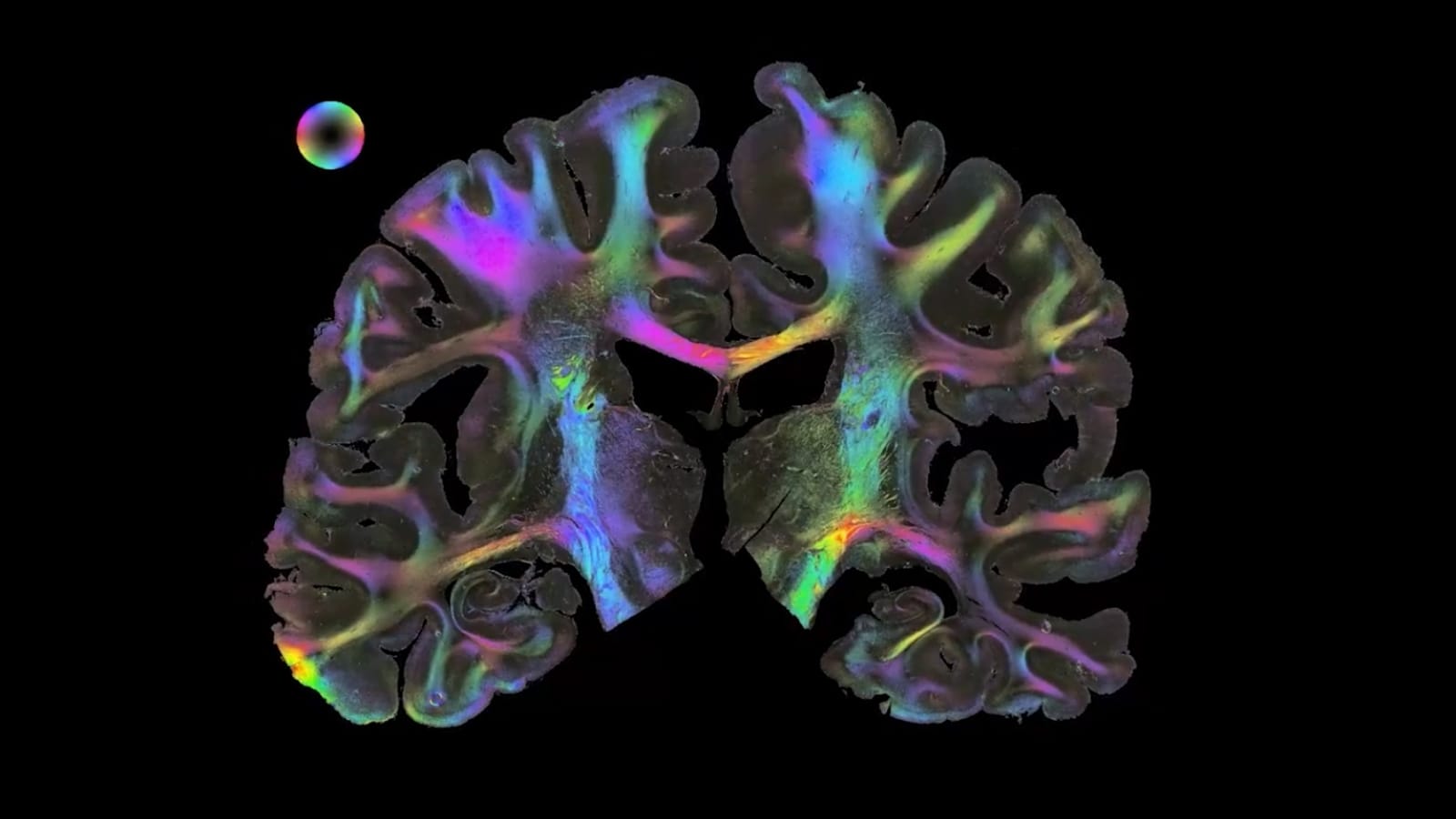
Fibre Architecture
Nerve fibres revealed by 3D-Polarized Light Imaging
Tap to learn more
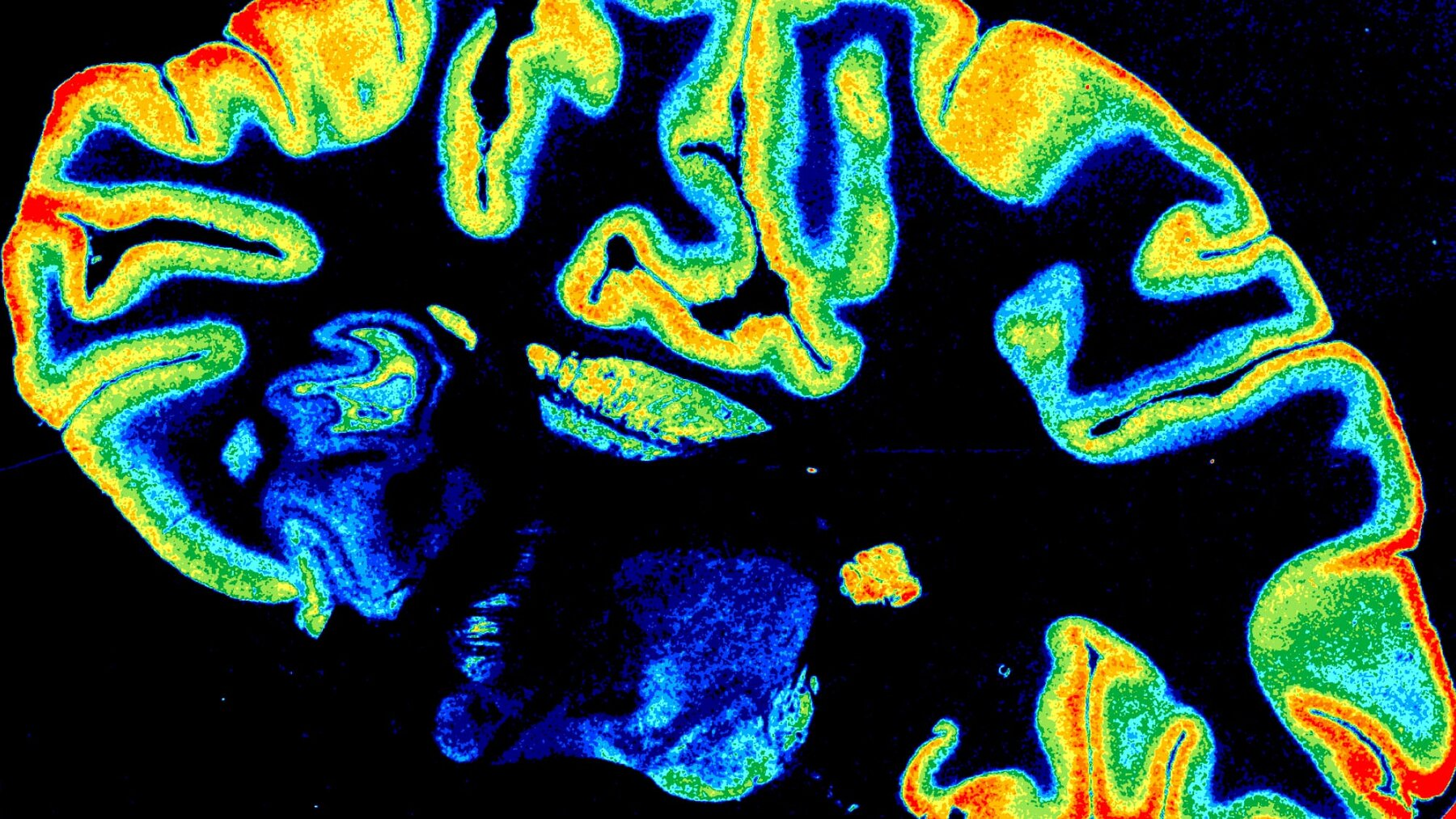
Neurotransmitter Receptors
Revealed by quantitative receptor autoradiography
Tap to learn more

Technology & Workflow
Atlas Technology
Building the Julich Brain Atlas involves imaging thousands of brain sections at high resolution producing data in the tera- to petabyte scale. Dealing with such massive amounts of data requires high-performance computing (HPC). To this end, we carry out research at the intersection of neuroinformatics, computer vision and artificial intelligence (AI).
Within the research group ‘Big Data Analytics’ led by Dr. Timo Dickscheid, we develop the digital methods and software tools required to acquire, process, analyse and store big image data and to build the three-dimensional, high-resolution atlas. We also coordinate the development of the brain atlas services of the digital infrastructure EBRAINS, which is built by the Human Brain Project (HBP) and provides open access to the Julich Brain Atlas.
High-Throughput Microscopy
To build the Julich Brain Atlas, we digitize whole-brain sections at high throughput. To deal with the large amounts of data produced, we develop distributed data management workflows that involve transfer of images to storage resources at the Jülich Supercomputing Centre (JSC).
Data Analytics with HPC
To generate maps and extract high-resolution features from a complete series of whole-brain sections at high throughput, we develop machine learning and computer vision algorithms. These help us to process and analyze the images in a mostly automated way at large scale.
Online viewers and programmatic software interfaces
Due to its large size, working with the atlas requires cloud technologies for remote access, visualization and annotation of imaging data. We develop the necessary software and metadata standards. Within the HBP, we have developed an interactive 3D viewer for browsing brain atlases at microscopic scale (“siibra-explorer”), a Python library for programmatic interaction with the atlas (“siibra-python”), and other web services.

Tools & Applications
Multilevel Human Brain Atlas of the HBP on EBRAINS
The Julich Brain Atlas forms the centrepiece of the open access Multilevel Human Brain Atlas of the digital research infrastructure EBRAINS, which has been built by the Human Brain Project (HBP).
Python interface for interacting with brain atlases
We have developed siibra-python, a Python client that allows the use of the Multilevel Human Brain Atlas in a programmatic and reproducible fashion. The client integrates brain parcellations and reference spaces across different spatial scales, connects them with multimodal regional data features, and provides many ways to use them.
EBRAINS Knowledge Graph
The data of the Julich Brain Atlas can be found on the EBRAINS Knowledge Graph, for which we have developed the comprehensive metadata standard openMINDS. By hosting a team of data curators, we also provide practical community support for integrating and sharing data via EBRAINS according to the FAIR principles.
Julich-Brain Gene Expression Tool (JuGEx)
The JuGEx tool enables the analysis of gene expressions in different cytoarchitectonic brain areas by integrating Julich-Brain and the Allen Human Brain Atlas, which is a rich resource of regional gene expression data. JuGEx integrates tissue transcriptome and cytoarchitectonic segregation and provides a framework for statistical analysis of differential gene expression. The JuGEx algorithm is available in the form of its original Matlab implementation and has been reimplemented as an extension to siibra-python as well as an interactive plugin for the atlas viewer “siibra-explorer”.
Julich-Brain Anatomy Toolbox
The Julich-Brain Anatomy Toolbox (a.k.a. SPM Anatomy Toolbox) is an SPM plugin with the probabilistic cytoarchitectonic maps of the Julich-Brain for combination with functional neuroimaging data.