BigBrain
The BigBrain
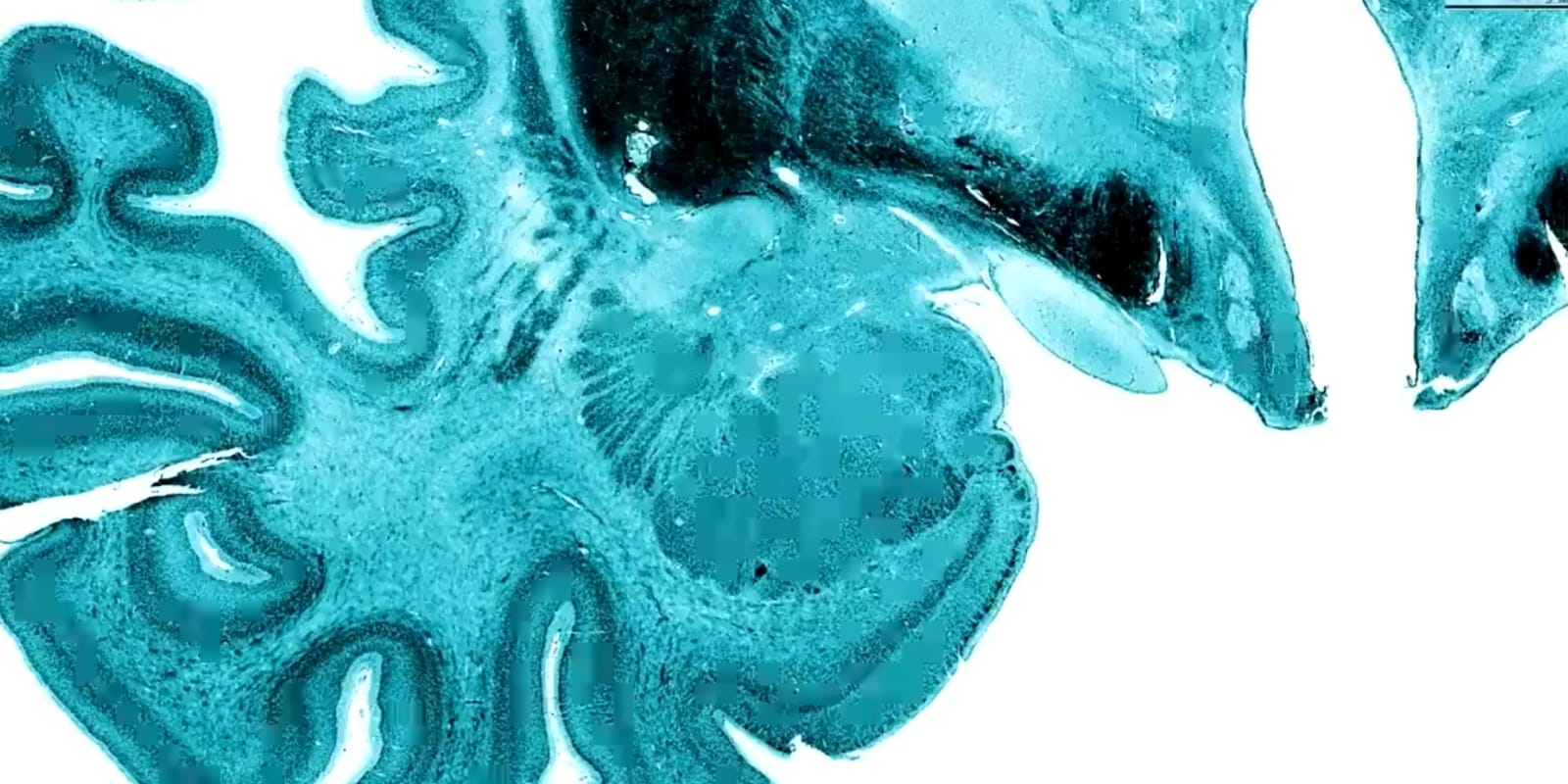
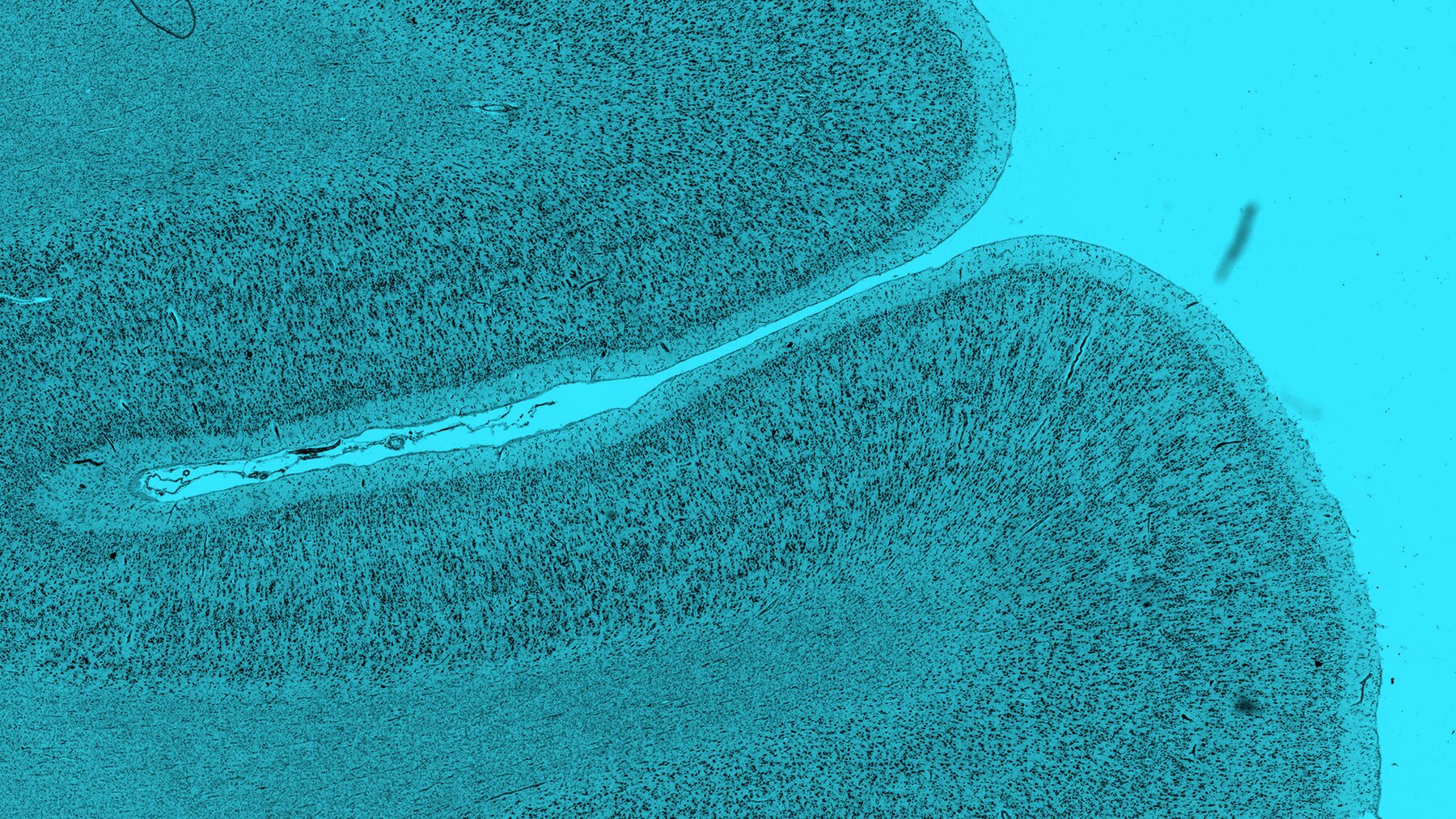
BigBrain is a three-dimensional reconstruction of an individual human brain stained for cell bodies at ultra-high, nearly cellular, resolution. BigBrain is unique in its scale and an invaluable tool for brain mapping: it enables the spatial anchoring of multimodal data into an anatomically realistic standard space in a functionally relevant, microscoscopic dimension. BigBrain is also one of ten individual brains whose cytoarchitectonic maps contribute to the Julich Brain Atlas.
BigBrain has been developed within a collaboration between Forschungszentrum Jülich and McGill University in Montreal.
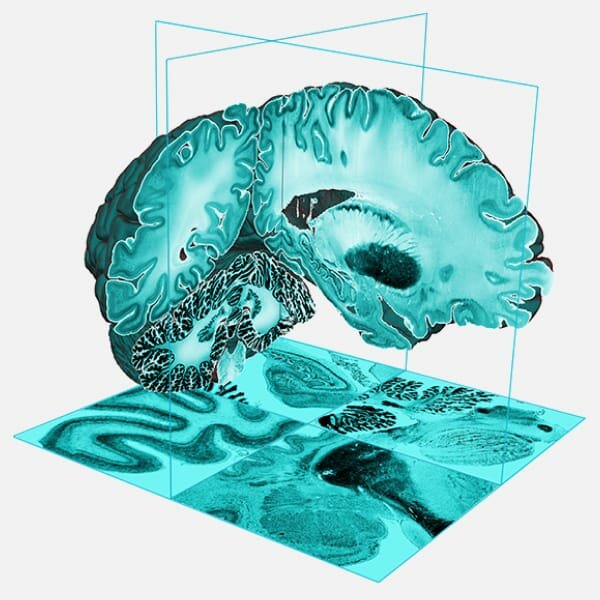
HIBALL
The German-Canadian Helmholtz International Lab HIBALL is a collaboration of neuroscientists and AI experts around Alan Evans at McGill University in Canada and Katrin Amunts at Forschungszentrum Jülich in Germany.
Within HIBALL, the next-generation multimodal 3D brain models will be developed at unmatched spatial resolution using novel deep learning methods and the latest supercomputing architectures to analyze neuroscientific data in the petabyte range.

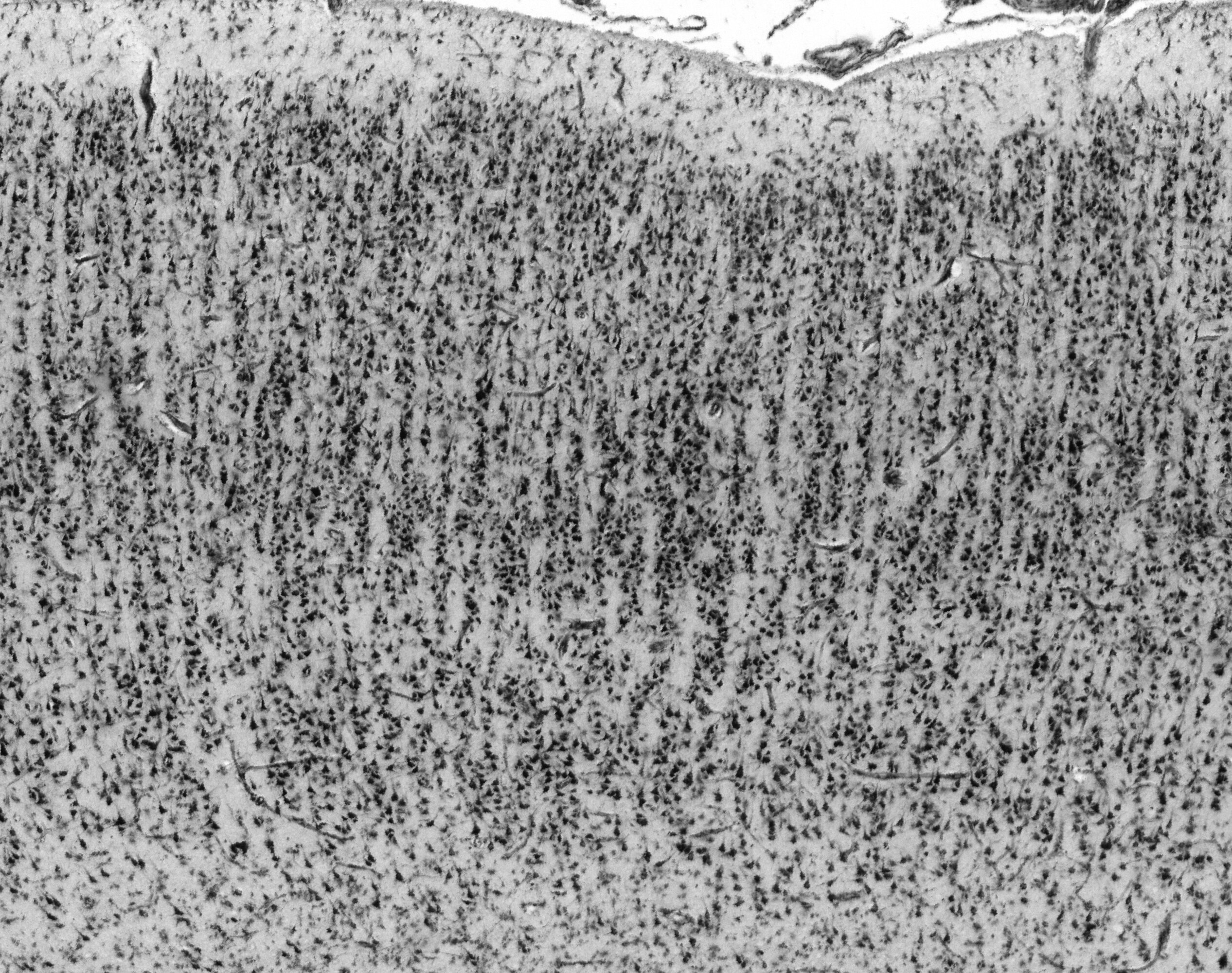
The Method in a Nutshell
To generate the BigBrain dataset, a complete paraffin-embedded human brain was cut into 7404 sections at 20-micrometer thickness and stained for cell bodies. The sections were digitized, resulting in images of up to 13,000 by 11,000 pixels and a data set of 1 TB. The images were downscaled to generate an isotropic resolution of 20 micrometers.
Applications & Data
BigBrain Dataset
The full dataset of images, volumes, and surfaces are available for download on an ftp site. Additional files can be accessed via LORIS.
EBRAINS Multilevel Human Brain Atlas
A three-dimensional atlas that integrates the different facets of human brain organisation at the millimeter and micrometer level
Cortical Layer Maps
Explore BigBrain cortical layer segmentation online.
Cytoarchitectonic Maps
Explore probabilistic cytoarchitectonic maps registered to the BigBrain online in the NeHuBa viewer.
Contact
Prof. Dr. med.
Katrin Amunts
Head of the research group Architecture and Brain Function
Institute of Neurosciences and Medicine (INM-1)
Forschungszentrum Jülich
D-52425 Jülich
Germany
Tel.: +49-2461-61-4300
Fax: +49-2461-61-3483
C. u. O. Vogt-Institute for Brain Research
Medical Faculty
University Hospital Düsseldorf
Life Science Center
Merowingerplatz 1A
D-40225 Düsseldorf
Germany
Tel: +49-211-81-06102
Fax: +49-211-81-06154

Key Publications
Convolutional Neural Networks for cytoarchitectonic brain mapping at large scale
NeuroImage 240: 118327 (2021), ISSN 1053-8119, doi
The BigBrainWarp toolbox for integration of BigBrain 3D histology with multimodal neuroimaging
eLife 10:e70119
Deep learning networks reflect cytoarchitectonic features used in brain mapping
Scientific Reports 10, 22039
BigBrain 3D atlas of cortical layers: Cortical and laminar thickness gradients diverge in sensory and motor cortices
PlosBiology 18(4): e3000678
Hippocampal subfields revealed through unfolding and unsupervised clustering of laminar and morphological features in 3D BigBrain
NeuroImage 206(116328)
IO Challenges for Human Brain Atlasing Using Deep Learning Methods - An In-Depth Analysis
27th Euromicro International Conference on Parallel, Distributed and Network-Based Processing (PDP): 291-298
BigBrain: An ultrahigh-resolution 3D human brain model
Science, 340(6139): 1472-1475