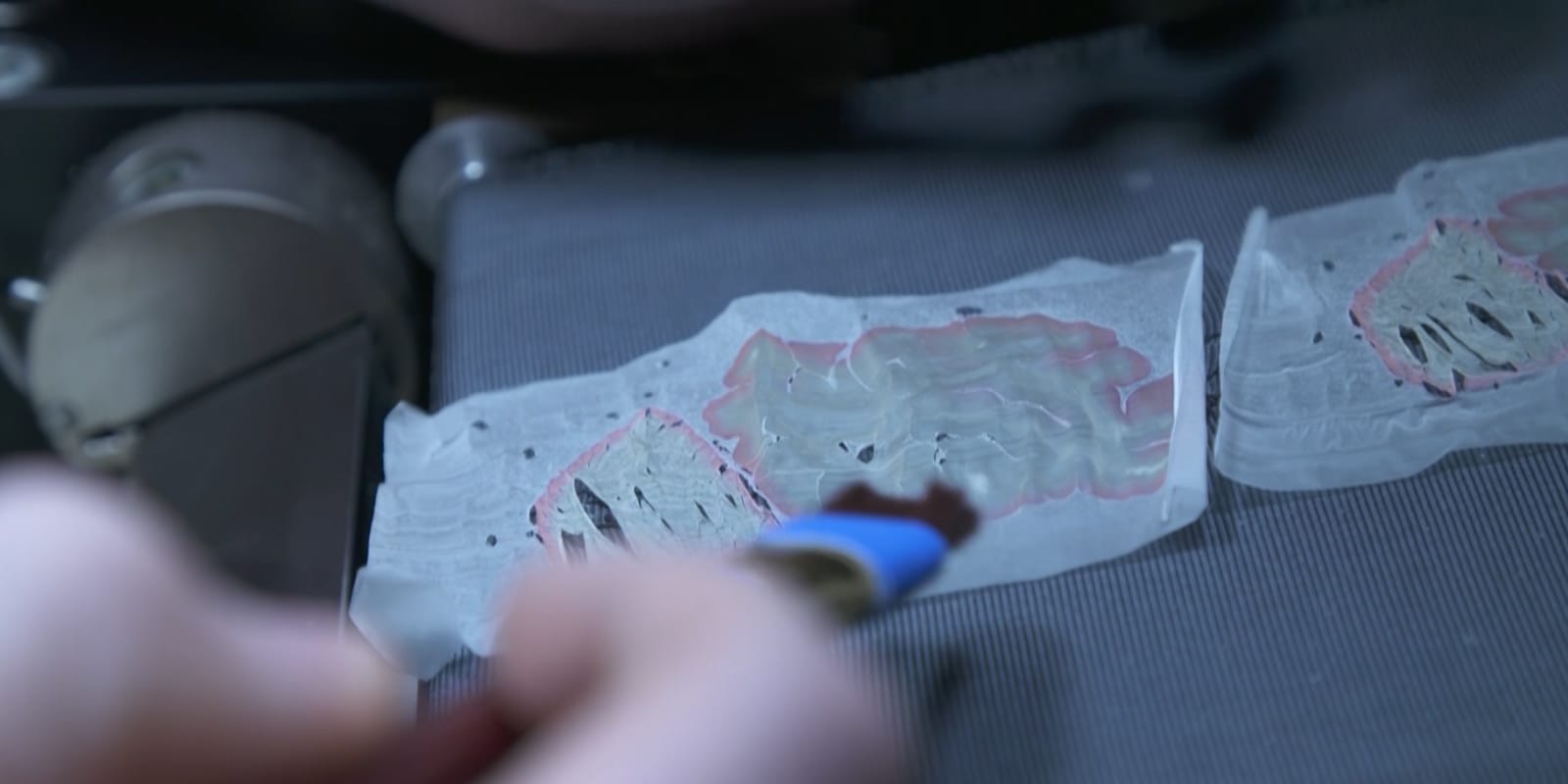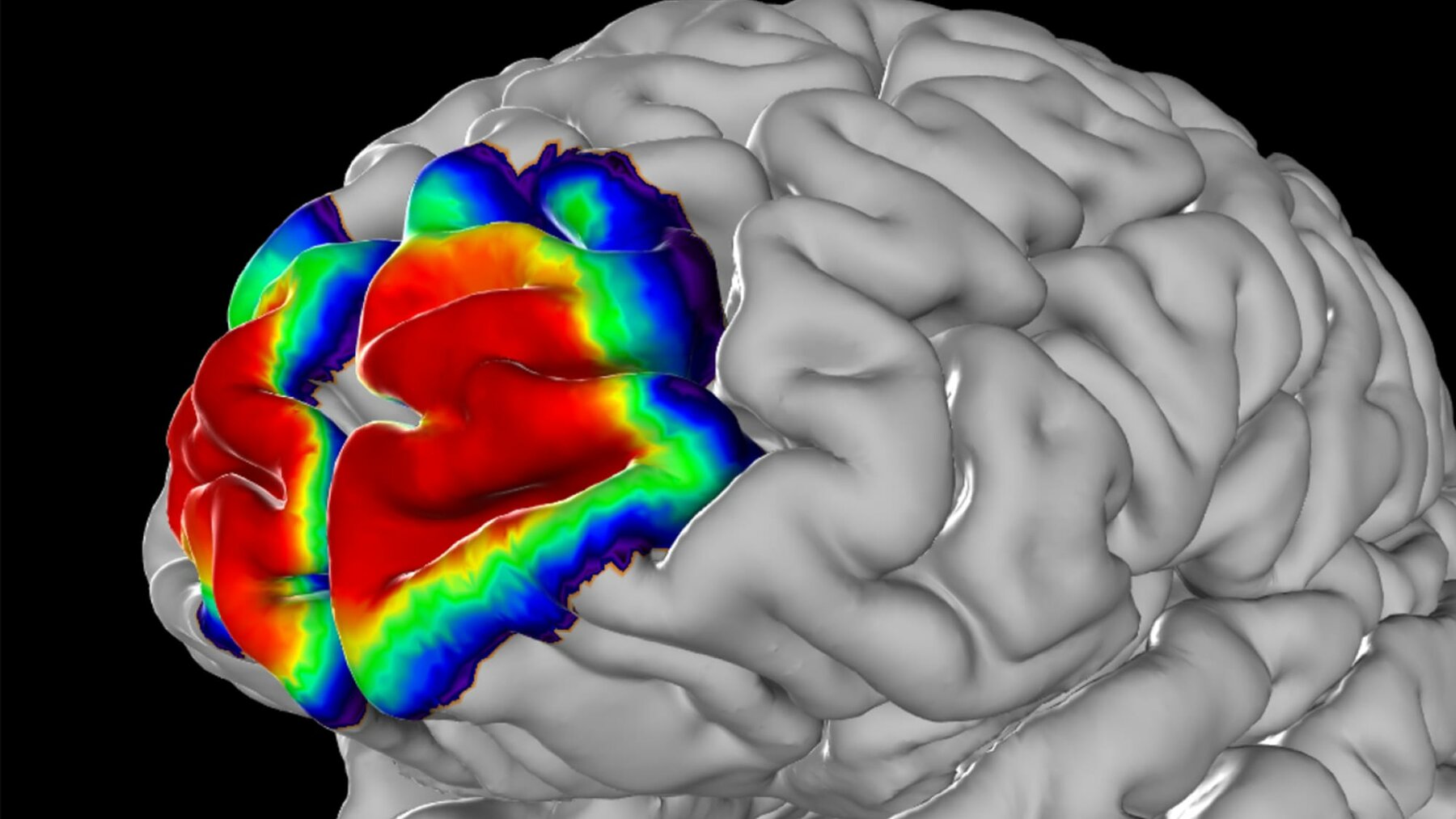
Probabilistic Maps
Cytoarchitectonic Probabilistic Maps in 3D Space
Julich-Brain provides maps of more than 200 brain areas based on their differences in cytoarchitecture, meaning the distribution, density and morphology of cells in a three-dimensional space. It considers variations between brains in their anatomy. Therefore, maps are probabilistic and reflect the interindividual variability in size and localisation of the areas in a three-dimensional space.
The Julich-Brain is the centrepiece of the Julich Brain Atlas as it allows to bring together and spatially align knowledge about the very distinct facets of the brain on a cellular level.
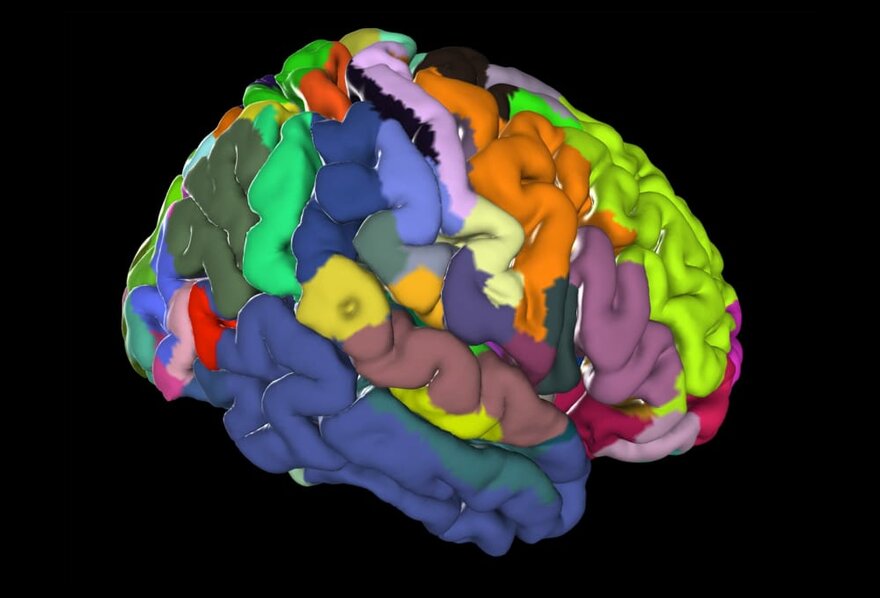
The Method in a Nutshell
The Julich-Brain is based on 23 post mortem brains from the body donor programme at the University of Düsseldorf. Brains were subjected to MRI and cut into 20 μm thick sections which were stained for cell bodies before digital imaging. Images of sections were 3D reconstructed, and aligned to a standard reference brain.
Borders between cytoarchitectonic areas were identified in brain sections using image analysis and statistical tools. Ten brains, five male and five female were studied to map each area. The areas were then 3D reconstructed, and superimposed in the reference brain. This allowed us to compute probability maps which provide measures of variability in areal size and localisation. Within the probability maps, colours code for the probability that an area can be found at a particular position. For these maps, regions which are not yet mapped are represented by so-called gap maps.
Probability maps can be simplified to show all areas at a glance: for this purpose, maximum probability maps were generated. Each position in the reference brain was assigned to the area with highest probability.
Data & Applications
Multilevel Human Brain Atlas of the HBP on EBRAINS
The Julich Brain Atlas forms the centrepiece of the open access Multilevel Human Brain Atlas of the digital research infrastructure EBRAINS, which has been built by the Human Brain Project (HBP).
Julich-Brain Anatomy Toolbox
The Julich-Brain Anatomy Toolbox (a.k.a. SPM Anatomy Toolbox) is an SPM plugin with the probabilistic cytoarchitectonic maps of the Julich-Brain for combination with functional neuroimaging data.
Julich-Brain Gene Expression Tool (JuGEx)
It enables analyses of gene expression with respect to a cytoarchitectonic brain area. The Allen Human Brain Atlas is a rich resource of regional gene expression data, and JuGEx allows to bring both worlds together: it integrates tissue transcriptome and cytoarchitectonic segregation and provides a framework for statistical analysis of differential gene expression in the adult human brain.
Contact
Prof. Dr. med.
Katrin Amunts
Head of the research group Architecture and Brain Function
Institute of Neurosciences and Medicine (INM-1)
Forschungszentrum Jülich
52425 Jülich
Germany
Tel.: +49-2461-61-4300
Fax: +49-2461-61-3483
C. u. O. Vogt-Institute for Brain Research
Medical Faculty
University Hospital Düsseldorf
Life Science Center
Merowingerplatz 1A
D-40225 Düsseldorf
Germany
Tel: +49-211-81-06102
Fax: +49-211-81-06154

Key Publications
Brain research challenges supercomputing
Science 374: 1054-1055
Julich-Brain: A 3D probabilistic atlas of the human brain's cytoarchitecture
Science 369: 988-992
Integration of transcriptomic and cytoarchitectonic data implicates a role for MAOA and TAC1 in the limbic-cortical network.
Brain Structure and Function 223: 2335–2342
Architectonic Mapping of the Human Brain beyond Brodmann
Neuron 88: 1086-1107
Cytoarchitecture of the cerebral cortex--more than localization
Neuroimage 37: 1061-1065
A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data
NeuroImage 25: 1325-1335