Fibre Architecture
Structural Connectivity and Fibre Architecture
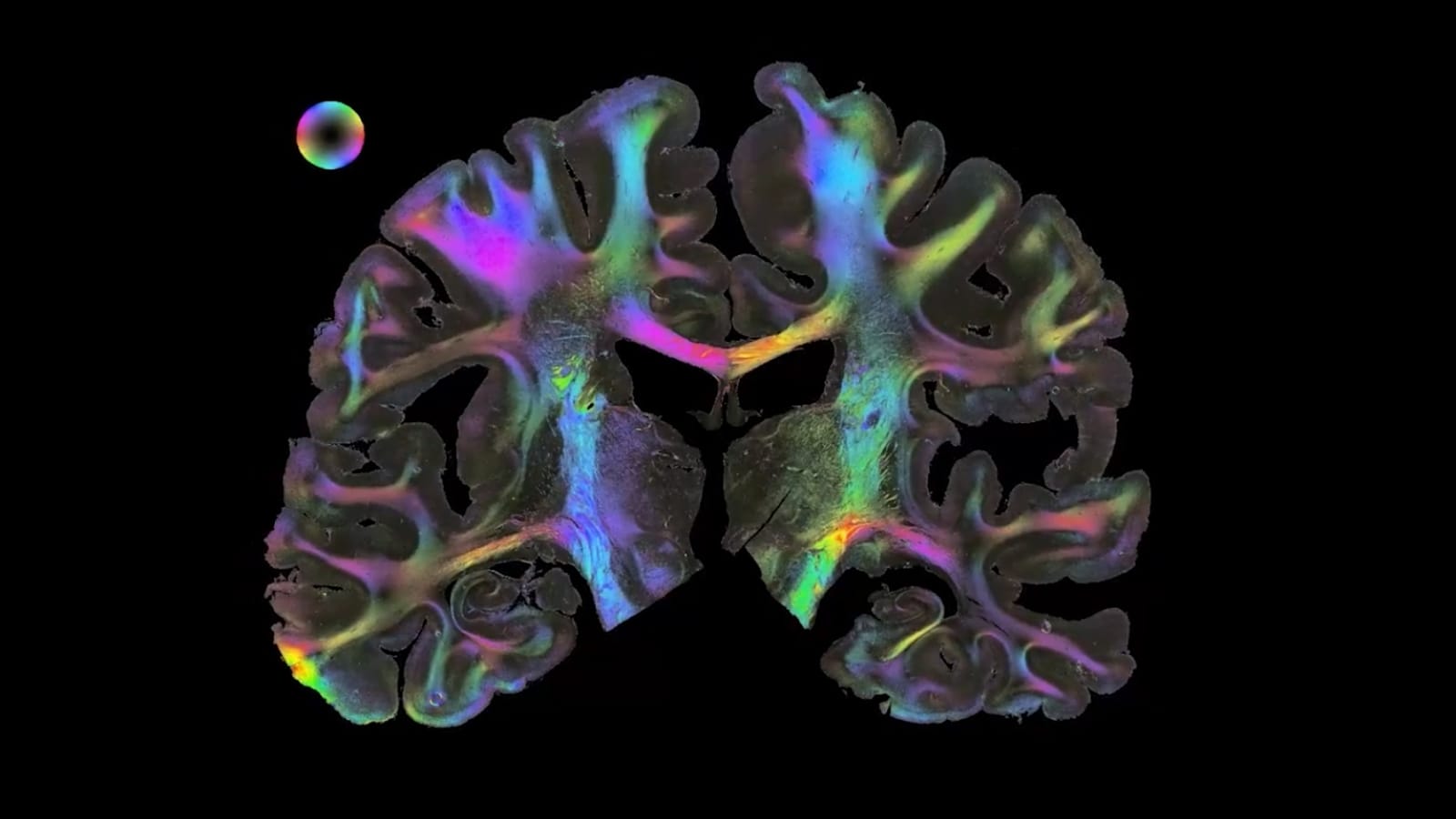
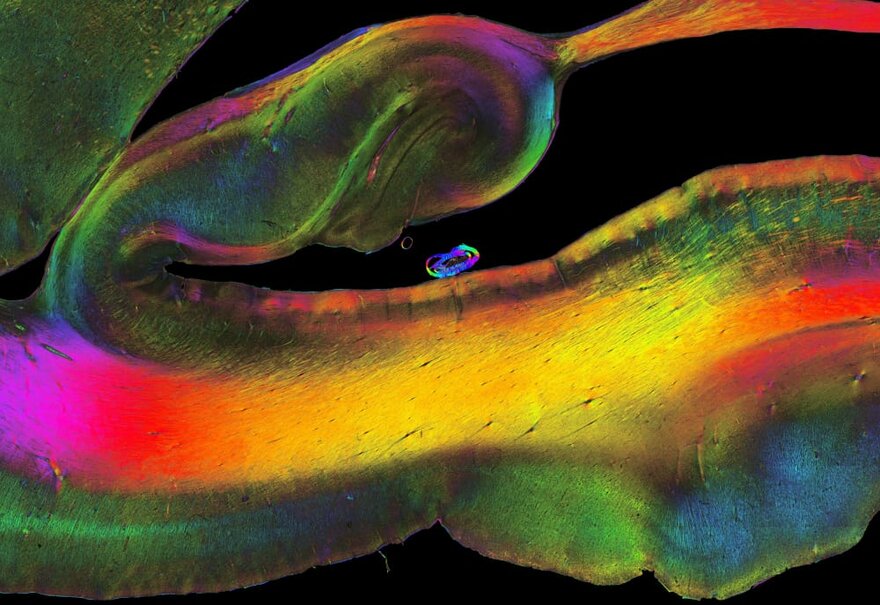
Three-dimensional polarized light imaging (3D PLI) allows us to visualise and analyse the architecture and connections of nerve fibres in post mortem brains at high resolution. It provides essential details about how information travels between different regions of the brain. We contrast distinct fibres and tracts and study their three-dimensional courses across serial brain sections with the aim of developing a 3D fibre atlas of the human brain. We are currently reconstructing brain regions at microscopic resolution, which is associated with large amounts of data. A first dataset of a reconstructed human hippocampus has been integrated into the EBRAINS Human Brain Atlas.
3D Polarised Light Imaging at a glance. Circularly polarized light is passed through an unstained histological human brain section, where its polarisation state is altered by the interaction with birefringent nerve fibres. This alteration becomes measurable by means of a rotating polarizer in front of a camera (left) and leads to changes in the sensed light intensities which are strongly correlated with the spatial orientation of fiber structures (middle). Extracted fiber orientations are finally assigned with colors and build the fiber orientation map (right).
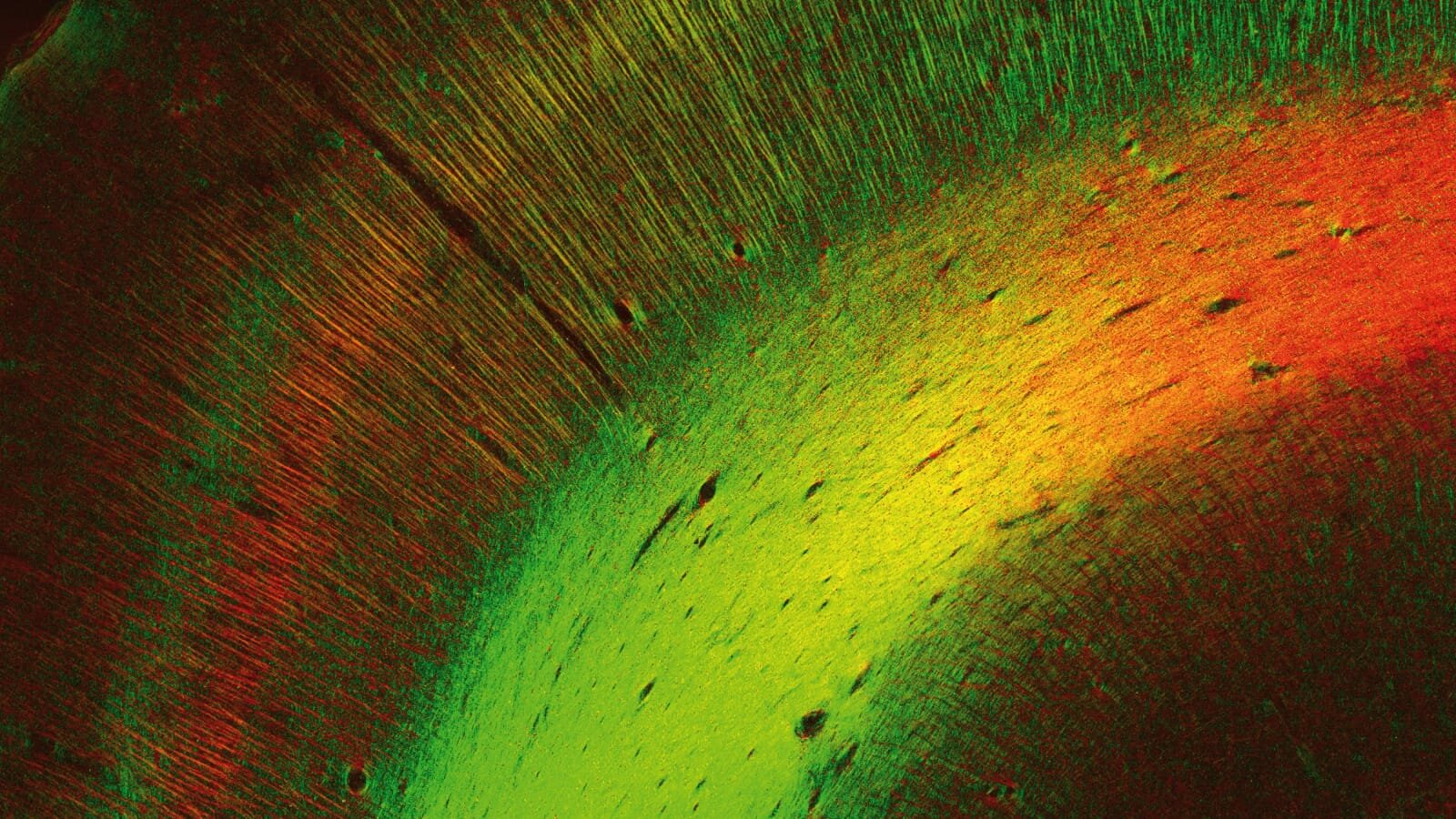
Polarized Light Imaging, © Mareen Fischinger

The method in a nutshell
Polarized light imaging makes use of birefringence, an optical effect of myelinated nerve fibres which causes a change of the oscillation behaviour (i.e., the polarization state) of an interacting light wave. To achieve high-quality measurements, the integrity of the myelin sheaths needs to be preserved. This requires specific tissue handling and preparation. To preserve myelin sheaths, conserved brain tissue is sectioned at very low temperatures (-50 °C).
During sectioning, en face images of the remaining brain block are acquired (blockface imaging) serving as reference images for later reconstruction of the initial brain shape. The generated unstained histological brain sections are scanned with different types of polarimetric setups, which are generally speaking composed of one linear and one circular polarizer and a green-wavelength light source. High-performance computing and simulation approaches enable reliable interpretation and visualization of the targeted fibre architecture.
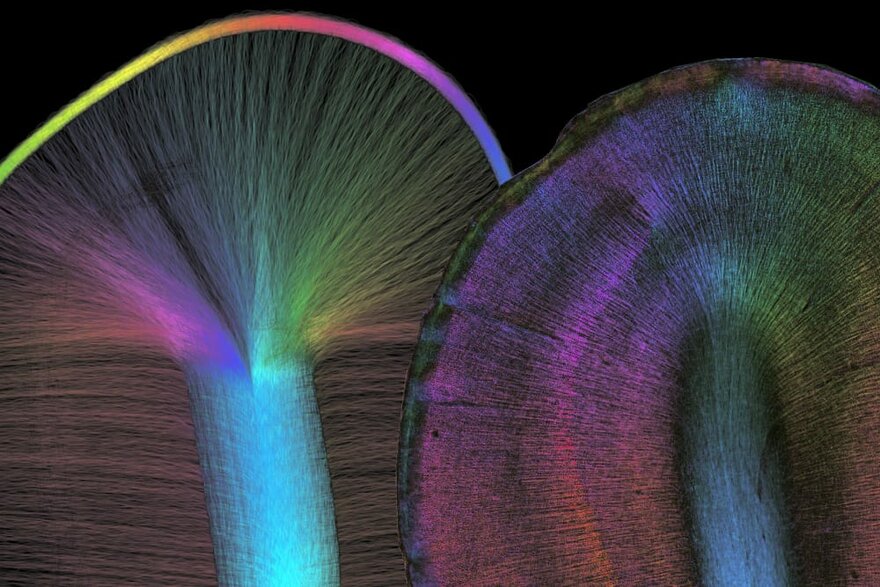
Combining Imaging Methods to Understand Connectivity
To understand the brain’s connectivity patterns over different scales, we also develop novel imaging techniques that study, for example, the scattering of light in brain tissue. Furthermore, we establish protocols and methods to combine 3D-PLI with complementary imaging methods, including Diffusion MRI, 3D-PLI, Two-Photon Fluorescence Microscopy, Mass Spectrometry Imaging and X-ray Diffraction.
Data & Applications
Several PLI datasets are openly available for download via the EBRAINS Knowledge Graph. Data on the human hippocampus has also been integrated into the EBRAINS Multilevel Human Brain Atlas.
Human Hippocampus Datasets
Datasets of fiber structures of a human hippocampus based on joint DMRI, 3D-PLI, and TPFM acquisitions are available on EBRAINS.
Contact
Prof. Dr.
Markus Axer
Head of Research Group Fibre Architecture
Institute of Neuroscience and Medicine (INM-1)
Forschungszentrum Jülich
52425 Jülich
Germany
Phone: +49 2461 61-6314
Email: m.axer@fz-juelich.de

Key Publications
Toward a High-Resolution Reconstruction of 3D Nerve Fiber Architectures and Crossings in the Brain Using Light Scattering Measurements and Finite-Difference Time-Domain Simulations
Physical review X 10(2), 021002
A cortex-like canonical circuit in the avian forebrain
Science 369(6511), eabc5534
Derivation of Fiber Orientations From Oblique Views Through Human Brain Sections in 3D-Polarized Light Imaging
Frontiers in neuroanatomy 12, 75
A novel approach to the human connectome: ultra-high resolution mapping of fiber tracts in the brain
NeuroImage 54, 1091 - 1101
High-resolution fiber tract reconstruction in the human brain by means of the three-dimensional polarized light imaging (3D-PLI)
Frontiers in Neuroinformatics 5, 1-13